Hamid Tizhoosh, PhD, Mayo Clinic

Photo by Dall-E
Foundation Models
The term foundation model (FM) in AI generally refers to large-scale (very deep) language models that can be utilized as a pre-trained and fine-tuned backbone (foundation) for many different applications. FMs are typically pre-trained on massive amounts of data to capture the intrinsic patterns, relationships, and representations of natural language (and increasingly images). FMs can be fine-tuned - after they have been pre-trained - for specific downstream tasks to deliver high-performance results on a wide range of applications.
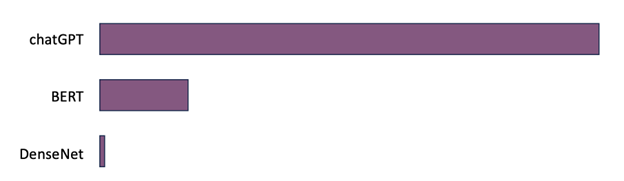
Figure 1. DenseNet with 7 million parameters is not a foundation model. The BERT model with 350 million parameters, and chatGPT with 1.5 billion parameters constitute foundation models (coarse lengths to conceptually visualize ratios).
FMs are exhibiting exceptional language understanding and generation capabilities. Some of the well-known FMs include OpenAI's GPT (Generative Pre-trained Transformer) series (such as GPT-3), Google's BERT (Bidirectional Encoder Representations from Transformers), and Facebook's RoBERTa (A Robustly Optimized BERT Pretraining Approach), among others [Wei2022].
Most FMs use the transformer architectures, which allows them to capture long-range dependencies in text through connectionist correlation analysis, making them highly effective for a wide variety of NLP tasks, including, translation, question-answering, and summarization. FMs can drastically reduce the need for extensive task-specific training data and computational resources if we employ transfer learning to exploit the knowledge learned, particularly through one-shot and few-shot learning.
FMs are becoming indispensable tools in dealing with text and other data modalities. However, there are also many ethical concerns attached to their training and usage. From impacting the employment landscape and environmental concerns (due to massive usage of computing resources) to bias and fairness, FMs may invoke controversial debates. For the medical field the questions of patient consent and privacy are crucial as well as the danger of fabrication and manipulation [Bommasani2021].
What do we need
Design and training FMs both pre-training and fine-tuning. During the massive pre-training, FMs learn from huge amounts of unlabeled data. The subsequent fine-tuning helps the model to adjust to specific downstream tasks using labeled data. For any conversational model, a large part of the FM must be based on the transformer architecture with layers of self-attention for discovering and exploiting long-range dependencies in the text. Without a transformer the true potentials of FMs for histopathology cannot be exploited.
One needs an insanely large and diverse corpus of data to train FMs. This commonly includes - for general purpose FMs - books, articles, websites, and social media posts, among others. Of course for histopathology, most of these sources may either be infeasible (e.g., copyright issues for medical books) or are not reliable (e.g., website and social media).
It is common knowledge that training a foundation model needs massive computational resources such as powerful GPUs or TPUs.
Creating input-output pairs for pre-training would be a required pre-processing, a task that is a bit harder for histopathology than for general-purpose FMs. Whereas fine-tuning data after pre-training may be easy for general-purpose FMs (e.g., through crowd-sourcing), this will be a real challenge in histopathology. The number of pathologists is low and the ones in clinical practice are in high-demand. Hospitals won’t be able to spare pathologists’ time for labeling data for FM fine-tunings. The model validation in histopathology cannot be performed with usual metrics; it has to be done live in clinical practice as a simultaneous research project. It would take months to properly validate a FM.
CLIP: The Role Model?
CLIP stands for "Contrastive Language-Image Pre-training", a FM developed by OpenAI that bridges natural language and digital images by learning “joint representations” of images and their corresponding textual descriptions. Based on availability of a large dataset containing images and their associated textual descriptions, CLIP learns to map similar images and their descriptions in a close proximity in the feature space while, at the same time, pushing dissimilar images and descriptions farther apart.
CLIP can perform “Image-to-Text Matching” when for any given image it predicts the most relevant description from a set of candidate descriptions. CLIP is bidirectional and can perform “Text-to-Image Matching” as well; Given a description, CLIP predicts the most relevant image from a set of candidate images. Various vision-related tasks can be performed with few-shot learning, where it can recognize and generate descriptions for images without extensive task-specific training.
CLIP has demonstrated impressive capabilities across a range of tasks, such as image classification, object detection, and even generating images from textual descriptions. Its ability to generalize across diverse datasets and tasks has made it a versatile and powerful tool in the field of AI research and applications. However, CLIP does not offer context-based conversations like chatGBPT.
Requirements for a FM in Histopathology
GPT-3 has 175 billion weights (300-500 petaFLOPS) and has used 4096 GPUs to be trained on hundreds of Terabytes of text data. And CLIP, to come closer to histopathology, used 400,000,000 image-caption pairs for its training. These numbers should guide us in the right direction of what data volume may be needed for us in histopathology to train a FM.
In computer science we talk about "Garbage In, Garbage Out", a fundamental principle, short GIGO, that emphasizes the importance of data quality in producing meaningful and accurate output from algorithms and models. GIGO basically states that if we provide incorrect or low-quality data as input to a computer program or system, the resulting output will also be incorrect or of low quality. In other words, the quality of the output is entirely dependent on the quality of the input. GIGO means that even the most sophisticated AI models will not produce reliable results if trained with low-quality data (inaccurate or irrelevant).
The quality of general (non-medical) text and images may be easily verifiable. The quality verification of the photo of a dog and its description “a dog plays in the park” does not pose an insurmountable challenge. However, in medicine both images and their descriptions are much more complex. The computerized quality control of medical data (meaning, association and relevance) is practically an unsolved problem that needs expert intervention. CLIP and other FMs may be able to get their data from the Internet but this is not a sane approach in medicine. Scraping histopathology data from the Internet is a clumsy approach that may harbor serious dangers for downstream tasks. We should avoid working with Internet data for medicine, even for research.
In histopathology we deal with whole slide images (WSIs). These are gigapixel color images that can easily be 100,000 by 100,000 pixels. Assuming every WSI has only one relevant region (i.e., abnormality or malignancy) that matches the diagnostic report or the molecular data, and assuming that “patching” (Figure 2) can serve as augmentation (many patches for the same text/caption), then we may be able to estimate how many WSIs we need to properly train a foundation model for histopathology. The two stage k-means clustering proposed by the search engine Yottixel provides a “mosaic” of an average of 80 patches, each approximately 1000 by 1000 pixels [Kalra2020].
Considering these facts we should also recall that the proper training of CLIP took 400,000,000 image-caption pairs [Radford2021]. That means to do something comparable in histopathology we would need 5,000,000 WSIs and their corresponding reports. This must be supplemented with corresponding lab data, radiology, genomics (DNA, RNA), social determinants and any other data type attached/related to those WSIs. It will be a monumental data management project to keep WSIs in the same place but perhaps pull the other data during the training.
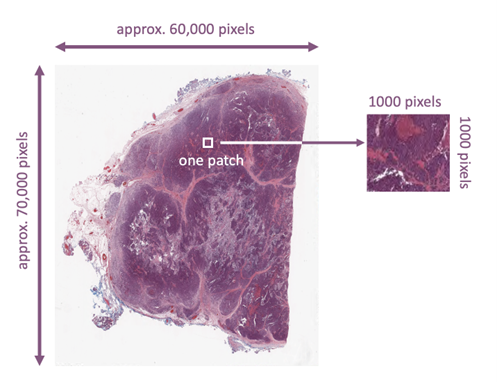
Figure 2. A whole slide image (WSI) is a large image containing many patches.
To this date, the largest public WSI dataset (namely TCGA) has barely 40,000 mixed WSIs (frozen and paraffin-embedded) with sparse textual descriptions. Scraping data from PubMed and other heterogeneous datasets to find image-text pairs relevant to histopathology will be working at the event horizon of the GIGO blackhole. The field of medicine needs high-quality clinical data to train FMs for pathology, radiology or any other subfield. That takes time. Serious researchers should exercise patience and put things together for the histopathology moonshot: train a foundation model with high-quality data from one or more hospitals.
References
[Radford2021] Radford, Alec, Jong Wook Kim, Chris Hallacy, Aditya Ramesh, Gabriel Goh, Sandhini Agarwal, Girish Sastry et al. "Learning transferable visual models from natural language supervision." In International conference on machine learning, pp. 8748-8763. PMLR, 2021.
[Kalra2020] Kalra, Shivam, Hamid R. Tizhoosh, Sultaan Shah, Charles Choi, Savvas Damaskinos, Amir Safarpoor, Sobhan Shafiei et al. "Pan-cancer diagnostic consensus through searching archival histopathology images using artificial intelligence." NPJ digital medicine 3, no. 1 (2020): 31.
[Wei2022] Wei, Jason, Yi Tay, Rishi Bommasani, Colin Raffel, Barret Zoph, Sebastian Borgeaud, Dani Yogatama et al. "Emergent abilities of large language models." arXiv preprint arXiv:2206.07682 (2022)
[Bommasani2021] Bommasani, Rishi, Drew A. Hudson, Ehsan Adeli, Russ Altman, Simran Arora, Sydney von Arx, Michael S. Bernstein et al. "On the opportunities and risks of foundation models." arXiv preprint arXiv:2108.07258 (2021).
Disclaimer: In seeking to foster discourse on a wide array of ideas, the Digital Pathology Association believes that it is important to share a range of prominent industry viewpoints. This article does not necessarily express the viewpoints of the DPA; however, we view this as a valuable point with which to facilitate discussion.
1 comment(s) on "The Pathology Moonshot - We need 5 Million WSIs to Train a Foundation Model"
09/29/2023 at 05:56 AM
Esther Abels says:
You mention "Serious researchers should exercise patience and put things together for the histopathology moonshot: train a foundation model with high-quality data from one or more hospitals." Agreed. It takes time to build it. It is up to us to do so. AI will be smarter than us, we have to ensure that it will also support us to be better. Our next generation should not have to wonder whether the data is reliable and accurate. Data collection and sharing is key. Let us make this a priority together, to be successful together. For the patient.Please log in to your DPA profile to submit comments